Software
Some software tools I developed during my PhD to facilitate easier and more reproducible analysis of chemical data.
|
chromatographR : Chromatographic Data Analysis Toolset
Github
|

|
|
|
|
|
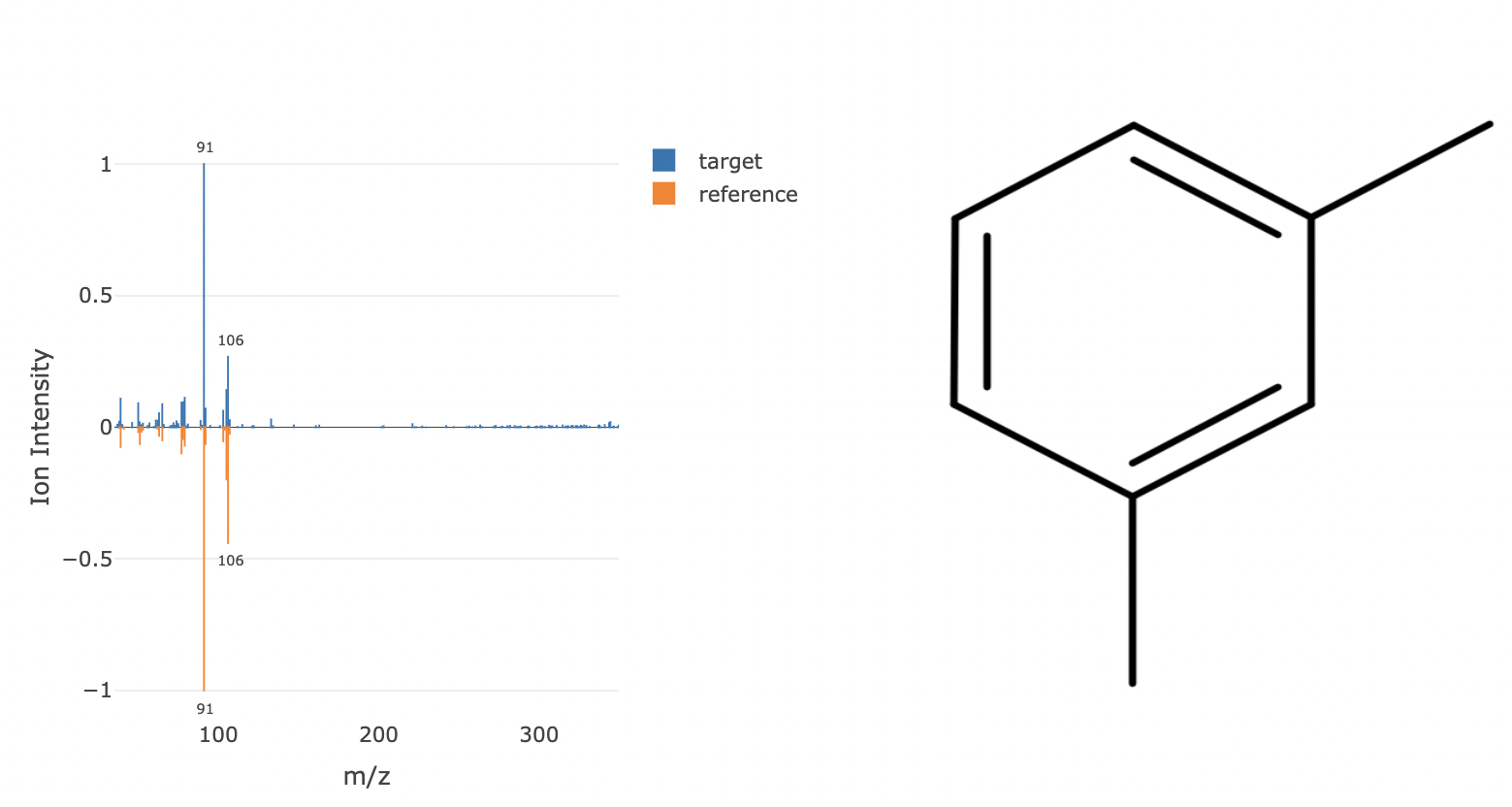
mzinspectr: Read and Analyze Mass Spectrometry Alignment Files .
Github
|
|
|
|
|
|
zerenebatchR: Utility for Batch Processing Images in Zerene Stacker .
Github
|

|
|
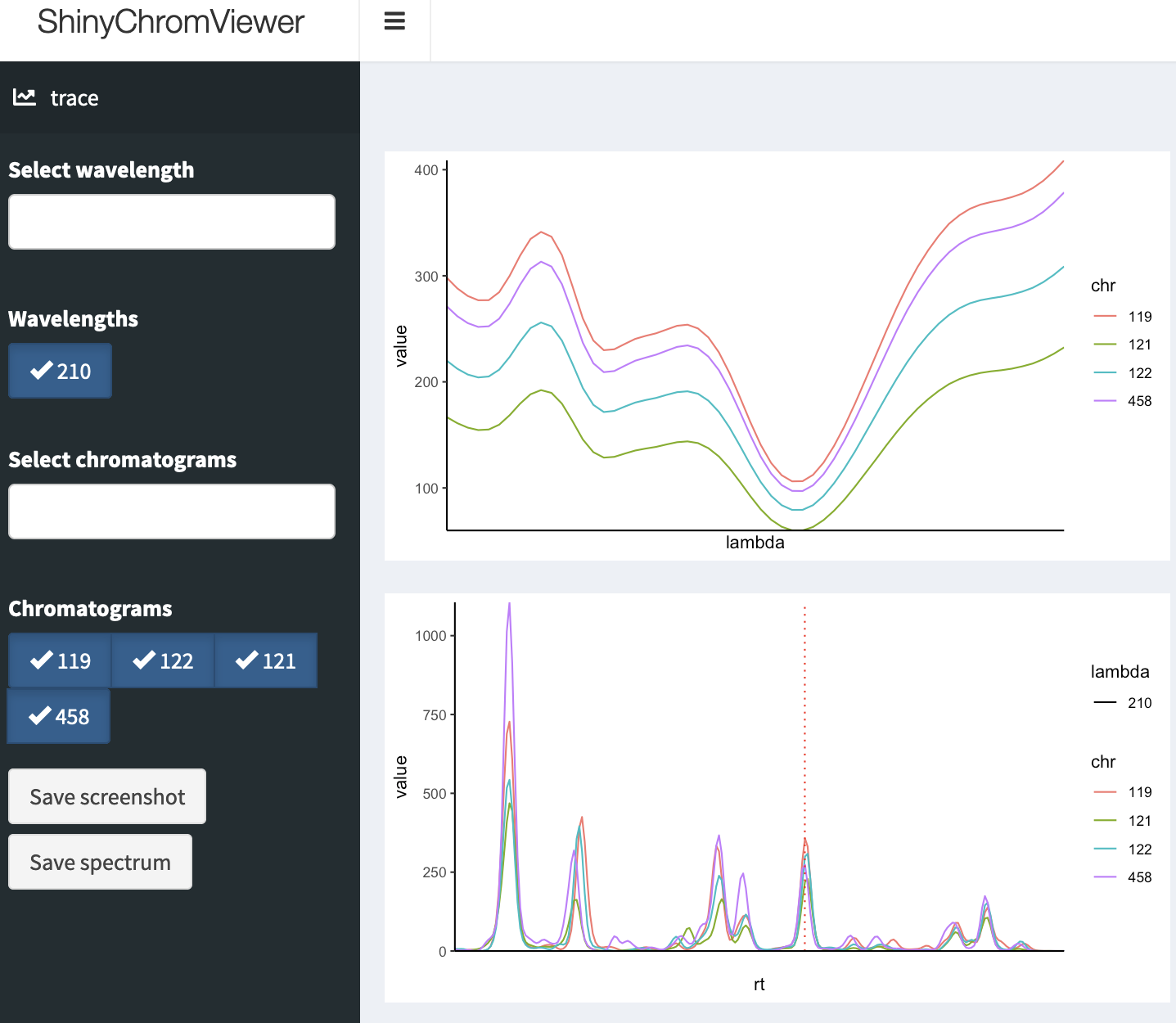
ShinyChromViewer: Shiny Gadget for Interactive Viewing and Exploration of Chromatograms .
Github
|
|
|
r2zotero: Send Suggested R Package Citations Directly to Zotero .
Github
|
Maintainer
VPdtw: Variable Penalty Dynamic Time Warping .
Github

I am also the current maintainer of the VPdtw: Variable Penalty Dynamic Time Warping, originally developed by David Clifford and Glenn Stone . I rescued this package from the CRAN archive and now use it as one of the options in chromatographR for aligning chromatograms.
Contributor
I have also contributed code to several other open source software projects:
webchem: Chemical Information from the Web
Github
RaMS: R-based access to Mass-Spectrometry data
Entab: * -> TSV







